2022
|
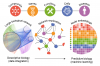
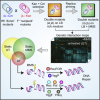
Integrating and formatting biomedical data as pre-calculated knowledge graph embeddings in the Bioteque,
Nature Communications,
2022,
DOI: https://doi.org/10.1038/s41467-022-33026-0
|
|
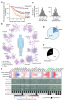
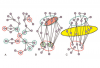
A proteome-scale map of the SARS-CoV-2-human contactome,
Nature Biotechnology,
2022, Pubmed: 36217029,
DOI: https://doi.org/10.1038/s41587-022-01475-z
|
|
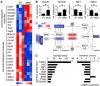
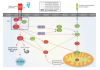
A community challenge for a pancancer drug mechanism of action inference from perturbational profile data,
Cell Reports Medicine,
2022, 3, Pubmed: 35106508,
DOI: 10.1016/j.xcrm.2021.100492
|
|
Detection of Nuclear Biomarkers for Chromosomal Instability,
Methods in Molecular Biology,
2022, 2445:117-125, Pubmed: 34972989,
DOI: 10.1007/978-1-0716-2071-7_8
|
|
Connecting chemistry and biology through molecular descriptors,
Current Opinion in Chemical Biology,
2022, 66:10290, Pubmed: 34626922,
DOI: 10.1016/j.cbpa.2021.09.001
|
2021
|
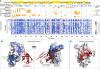
Binary Interactome Models of Inner-Versus Outer-Complexome Organization,
bioRxiv,
2021,
DOI: 10.1101/2021.03.16.435663
|
|
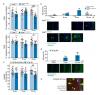
Identification and drug-induced reversion of molecular signatures of Alzheimers disease onset and progression in AppNL-GF, AppNL-F and 3xTg-AD mouse models,
Genome Medicine ,
2021, 13, Pubmed: 34702310,
DOI: https://doi.org/10.1186/s13073-021-00983-y
|
|
Haste makes waste: A critical review of docking-based virtual screening in drug repurposing for SARS-CoV-2 main protease (M-pro) inhibition,
Medicinal research reviews,
2021, Pubmed: 34697818,
DOI: 10.1002/med.21862
|
|
Identification of Broad-Spectrum MMP Inhibitors by Virtual Screening,
Molecules,
2021, 26:4553, Pubmed: 34361703,
DOI: 10.3390/molecules26154553
|
|
Association of Transcriptomic Signatures of Inflammatory Response with Viral Control after Dendritic Cell-Based Therapeutic Vaccination in HIV-1 Infected Individuals,
Vaccines,
2021, 9:799, Pubmed: 34358215,
DOI: 10.3390/vaccines9070799
|
|
Bioactivity descriptors for uncharacterized compounds,
Nature Communications,
2021, 12:1-13, Pubmed: 34168145,
DOI: 10.1038/s41467-021-24150-4
|
|
Anti-Inflammatory and Immunomodulatory Effects of the Grifola frondosa Natural Compound o-Orsellinaldehyde on LPS-Challenged Murine Primary Glial Cells. Roles of NF-κβ and MAPK,
Pharmaceutics,
2021, 13:806, Pubmed: 34071571,
DOI: 10.3390/pharmaceutics13060806
|
|
Natural variants suppress mutations in hundreds of essential genes,
Molecular systems biology,
2021, 17:e10138, Pubmed: 34042294,
DOI: 10.15252/msb.202010138
|
|
Environmental robustness of the global yeast genetic interaction network,
Science,
2021, 372:6542, Pubmed: 33958448,
DOI: 10.1126/science.abf8424
|
|
Analysing the yeast complexome—the Complex Portal rising to the challenge,
Nucleic acids research,
2021, 49:3156-3167, Pubmed: 33677561,
DOI: /10.1093/nar/gkab077
|
2020
|
Systematic analysis of bypass suppression of essential genes,
Molecular Systems Biology,
2020, 16(9):e9828, Pubmed: 32939983,
DOI: 10.15252/msb.20209828
|
|
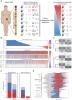
Personalized Cancer Therapy Prioritization Based on Driver Alteration Co-occurrence Patterns,
Genome Medicine,
2020, 12, Pubmed: 32907621,
DOI: https://doi.org/10.1186/s13073-020-00774-x
|
|
Personalized cancer therapy prioritization based on driver alteration co-occurrence patterns,
Genome Medicine,
2020, 12(1):78, Pubmed: 32907621,
DOI: 10.1186/s13073-020-00774-x
|
|
Bioactivity Profile Similarities to Expand the Repertoire of COVID-19 Drugs,
Journal of Chemical Information and Modeling,
2020, Pubmed: 32672454,
DOI: https://doi.org/10.1021/acs.jcim.0c00420
|
|
Publisher Correction: Extending the small-molecule similarity principle to all levels of biology with the Chemical Checker ,
10.1038/s41587-020-0564-6 ,
2020, 38(9):1098, Pubmed: 32440008,
DOI: 10.1038/s41587-020-0564-6
|
|
Extending the small-molecule similarity principle to all levels of biology with the Chemical Checker,
Nature Biotechnology,
2020, 38(9):1087-1096, Pubmed: 32440005,
DOI: https://doi.org/10.1038/s41587-020-0502-7,
Supplementary material
|
|
Antigen-stimulated PBMC transcriptional protective signatures for malaria immunization,
Science Translational Medicine,
2020, 12, Pubmed: 32404508,
DOI: 10.1126/scitranslmed.aay8924
|
|
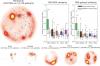
A reference map of the human binary protein interactome,
Nature ,
2020, 580:402-408, Pubmed: 32296183,
DOI: 10.1038/s41586-020-2188-x,
Supplementary material
|
|
Systematic genetics and single-cell imaging reveal widespread morphological pleiotropy and cell-to-cell variability,
Molecular Systems Biology,
2020, Pubmed: 32064787,
DOI: 10.15252/msb.20199243
|
|
A classifier to predict viral control after antiretroviral treatment interruption in chronic HIV-1 infected patients,
Journal of Acquire Immune Deficiency Syndrome,
2020, Pubmed: 31904703,
DOI: 10.1097/QAI.0000000000002281
|
2019
|
In silico Characterization of Human Prion-Like Proteins: Beyond Neurological Diseases,
Frontiers in Physiology,
2019,
DOI: https://doi.org/10.3389/fphys.2019.00314
|
|
DYRK1A Overexpression Alters Cognition and Neural-Related Proteomic Pathways in the Hippocampus That Are Rescued by Green Tea Extract and/or Environmental Enrichment,
Frontiers in molecular Neuroscience,
2019, Pubmed: 31803016,
DOI: 10.3389/fnmol.2019.00272
|
|
Truncating variant burden in high-functioning autism and pleiotropic effects of LRP1 across psychiatric phenotypes,
Journal of Psychiatry & Neuroscience,
2019, 44(5):350-359, Pubmed: 31094488,
DOI: https://doi.org/10.1503/jpn.180184
|
|
Encircling the regions of the pharmacogenomic landscape that determine drug response,
Genome Medicine,
2019, 11:17, Pubmed: 30914058,
DOI: https://doi.org/10.1186/s13073-019-0626-x
|
2018
|
Formatting biological big data for modern machine learning in drug discovery,
WIREs Computational Molecular Science,
2018,
DOI: 10.1002/wcms.1408
|
|
Cell-specific proteome analyses of human bone marrow reveal molecular features of age-dependent functional decline,
Nature Communications,
2018, 9(1):4004, Pubmed: 30275468,
DOI: 10.1038/s41467-018-06353-4
|
|
DynBench3D, a Web-Resource to Dynamically Generate Benchmark Sets of Large Heteromeric Protein Complexes,
Journal of Molecular Biology,
2018, 18:30769, Pubmed: 30274705,
DOI: https://doi.org/10.1016/j.jmb.2018.09.011
|
|
Exploring the OncoGenomic Landscape of cancer,
Genome Medicine,
2018, 10:61, Pubmed: 30071882,
DOI: 10.1186/s13073-018-0571-0
|
|
Systems analysis of intracellular pH vulnerabilities for cancer therapy,
Nature Communications,
2018, 9:2997, Pubmed: 30065243,
DOI: 10.1038/s41467-018-05261-x
|
|
The transcriptomic and epigenetic map of vascular quiescence in the continuous lung endothelium,
eLife,
2018, 7:e34423, Pubmed: 29749927,
DOI: 10.7554/eLife.34423
|
|
Rationalizing Drug Response in Cancer Cell Lines,
Journal of Molecular Biology,
2018, 430(18 Pt A):3016-3027, Pubmed: 29626539,
DOI: 10.1016/j.jmb.2018.03.021
|
|
Proteome data improves protein function prediction in the interactome of H. pylori,
Molecular & Cell Proteomics,
2018, 17(5):961-973, Pubmed: 29414760,
DOI: 10.1074/mcp.RA117.000474
|
2017
|
Drug repositioning beyond the low-hanging fruits,
Current Opinion in Systems Biology,
2017, 3:95–102,
DOI: 10.1016/j.coisb.2017.04.010
|
|
A framework for exhaustively mapping functional missense variants,
Molecular Systems Biology,
2017, 13(12):957, Pubmed: 29269382,
DOI: 10.15252/msb.20177908
|
|
Features of the Chaperone Cellular Network Revealed through Systematic Interaction Mapping,
Cell Reports,
2017, 20(11):2735-2748, Pubmed: 28903051,
DOI: http://dx.doi.org/10.1016/j.celrep.2017.08.074
|
|
Detecting similar binding pockets to enable systems polypharmacology,
PLoS Computational Biology,
2017, 13(6):e1005522, Pubmed: 28662117,
DOI: 10.1371/journal.pcbi.1005522
|
|
Mechanisms of suppression: The wiring of genetic resilience,
BioEssays,
2017, 39(7), Pubmed: 28582599,
DOI: 10.1002/bies.201700042
|
|
A PanorOmic view of personal cancer genomes,
Nucleic Acids Research,
2017, 45(W1):W195-W200, Pubmed: 28453651,
DOI: 10.1093/nar/gkx311
|
|
Quantification of pathway crosstalk reveals novel synergistic drug combinations for breast cancer,
Cancer Research,
2017, 77(2):459-469, Pubmed: 27879272,
DOI: 10.1158/0008-5472.CAN-16-0097
|
2016
|
Exploring genetic suppression interactions on a global scale,
Science,
2016, 354, Pubmed: 27811238,
DOI: 10.1126/science.aag0839
|
|
Cancer network activity associated with therapeutic response and synergism,
Genome Medicine,
2016, 8:88, Pubmed: 27553366,
DOI: 10.1186/s13073-016-0340-x,
Supplementary material
|
|
An inter‐species protein–protein interaction network across vast evolutionary distance,
Molecular Systems Biology,
2016, 12:865, Pubmed: 27107014,
DOI: 10.15252/msb.20156484
|
|
Pooled‐matrix protein interaction screens using Barcode Fusion Genetics,
Molecular Systems Biology,
2016, 12:863, Pubmed: 27107012,
DOI: 10.15252/msb.20156660
|
|
Risk assessment of Soulatrolide and Mammea (A/BA+A/BB) coumarins from Calophyllum brasiliense by a toxicogenomic and toxicological approach,
Food and Chemical Toxicology,
2016, 91:117-129, Pubmed: 26995226,
DOI: 10.1016/j.fct.2016.03.010
|
|
Widespread Expansion of Protein Interaction Capabilities by Alternative Splicing,
Cell,
2016, 164(4):805-817, Pubmed: 26871637,
DOI: 10.1016/j.cell.2016.01.029
|
|
Systems-Wide Prediction of Enzyme Promiscuity Reveals a New Underground Alternative Route for Pyridoxal 5’-Phosphate Production in E. coli,
PLOS Computational Biology,
2016, 12(1):1-19, Pubmed: 26821166,
DOI: 10.1371/journal.pcbi.1004705
|
|
Novel Neuroprotective Multicomponent Therapy for Amyotrophic Lateral Sclerosis Designed by Networked Systems,
PLoS One,
2016, 11(1):1-17, Pubmed: 26807587,
DOI: 10.1371/journal.pone.0147626
|
|
Conditional Epistatic Interaction Maps Reveal Global Functional Rewiring of Genome Integrity Pathways in Escherichia coli,
Cell Reports,
2016, 14:648-661, Pubmed: 26774489,
DOI: 10.1016/j.celrep.2015.12.060
|
|
Residues Coevolution Guides the Systematic Identification of Alternative Functional Conformations in Proteins,
Structure,
2016, 24:1-11, Pubmed: 26688214,
DOI: 10.1016/j.str.2015.10.025
|
2015
|
Isolation of Human Colon Stem Cells Using Surface Expression of PTK7,
Stem Cell Reports,
2015, 5:1-9, Pubmed: 26549850,
DOI: 10.1016/j.stemcr.2015.10.003
|
|
Drug sensitivity in cancer cell lines is not tissue-specific,
Molecular Cancer,
2015, 14:40:1-4, Pubmed: 25881072,
DOI: 10.1186/s12943-015-0312-6
|
|
Using neighborhood cohesiveness to infer interactions between protein domains,
Bioinformatics,
2015:1-8, Pubmed: 25838464,
DOI: 10.1093/bioinformatics/btv188
|
|
Network-based proteomic approaches reveal the neurodegenerative, neuroprotective and pain-related mechanisms involved after retrograde axonal damage,
Scientific Reports,
2015, 5: 9185:1-13, Pubmed: 25784190,
DOI: 10.1038/srep09185
|
|
dSysMap: Exploring the edgetic role of disease mutations,
Nature Methods,
2015, 12:167–168, Pubmed: 25719824,
DOI: 10.1038/nmeth.3289
|
|
Systematic identification of molecular links between core and candidate genes in breast cancer,
Journal of Molecular Biology,
2015, 427:1436-1450, Pubmed: 25640309,
DOI: 10.1016/j.jmb.2015.01.014
|
|
Breast Cancer Genes PSMC3IP and EPSTI1 Play a Role in Apoptosis Regulation,
PLoS ONE,
2015, 10(1): e0115352, Pubmed: 25590583,
DOI: 10.1371/journal.pone.0115352
|
|
Yeast Mitochondrial Protein-Protein Interactions Reveal Diverse Complexes and Disease-Relevant Functional Relationships,
Journal of Proteome Research, Advanced Online Publication,
2015, 14:1220−1237, Pubmed: 25546499,
DOI: 10.1021/pr501148q
|
|
IntSide: a web server for the chemical and biological examination of drug side effects,
Bioinformatics,
2015, 31 (4):612-613, Pubmed: 25380960,
DOI: 10.1093/bioinformatics/btu688
|
2014
|
The EHEC-host interactome reveals novel targets for the translocated intimin receptor,
Scientific Reports,
2014, 4: 7531, Pubmed: 25519916,
DOI: 10.1038/srep07531
|
|
A chemo-centric view of human health and disease,
Nature Communications,
2014, 5:5676, Pubmed: 25435099,
DOI: 10.1038/ncomms6676
|
|
A Proteome-Scale Map of the Human Interactome Network,
Cell,
2014, 159(5):1212–1226, Pubmed: 25416956,
DOI: 10.1016/j.cell.2014.10.050
|
|
Assessing the applicability of template-based protein docking in the twilight zone,
Structure,
2014, 22(9):1356–1362, Pubmed: 25156427,
DOI: 10.1016/j.str.2014.07.009
|
|
Increasing the precision of orthology-based complex prediction through network alignment,
PeerJ,
2014, 2:e413, Pubmed: 24918034,
DOI: 10.7717/peerj.413
|
|
An Second-generation Protein-Protein Interaction Network of Helicobacter pylori,
Molecular & Cellular Proteomics,
2014, 13:1318-1329, Pubmed: 24627523,
DOI: 10.1074/mcp.O113.033571
|
|
The binary protein-protein interaction landscape of Escherichia coli,
Nature Biotechnology,
2014, 32:285–290, Pubmed: 24561554,
DOI: doi:10.1038/nbt.2831
|
|
Charting the molecular links between driver and susceptibility genes in colorectal cancer,
Biochem. Biophys. Res. Commun,
2014, 445(4):734–738, Pubmed: 24412244,
DOI: 10.1016/j.bbrc.2013.12.012
|
|
3did: a catalogue of domain-based interactions of known three-dimensional structure,
Nucleic Acids Research,
2014, 42 (D1):D374-D379, Pubmed: 24081580,
DOI: 10.1093/nar/gkt887
|
2013
|
Towards a detailed atlas of protein-protein interactions,
Current Opinion in Structural Biology,
2013, 23(6):929–940, Pubmed: 23896349,
DOI: 10.1016/j.sbi.2013.07.005
|
|
Structural Systems Pharmacology: the role of 3D structures in next generation drug development,
Chemistry & Biology,
2013, 20(5):674-684, Pubmed: 23706634,
DOI: 10.1016/j.chembiol.2013.03.004
|
|
Functional and Structural Analysis of C-Terminal BRCA1 Missense Variants,
PLoS ONE,
2013, 8(4):e61302, Pubmed: 23613828,
DOI: 10.1371/journal.pone.0061302
|
|
Analysis of chemical and biological features yields mechanistic insights into drug side effects,
Chemistry & Biology,
2013, 20:594-603, Pubmed: 23601648,
DOI: 10.1016/j.chembiol.2013.03.017
|
|
The proton-pump inhibitor lansoprazole enhances amyloid beta production,
PLoS ONE,
2013, 8(3):e58837, Pubmed: 23520537,
DOI: 10.1371/journal.pone.0058837
|
|
Interactome3D: adding structural details to protein networks,
Nature Methods,
2013, 10(1):47-53, Pubmed: 23399932,
DOI: 10.1038/nmeth.2289,
Supplementary material
|
2012
|
NetAligner – A network alignment server to compare complexes, pathways and whole interactomes,
Nucleic Acids Res,
2012, Pubmed: 22618871,
DOI: 10.1093/nar/gks446
|
|
From protein interaction networks to novel therapeutic strategies,
IUBMB Life,
2012, Pubmed: 22573601,
DOI: 10.1002/iub.1040
|
|
Towards Alzheimer's root cause: ECSIT as an integrating hub between oxidative stress, inflammation and mitochondrial dysfunction,
BioEssays,
2012, Pubmed: 22513506,
DOI: 10.1002/bies.201100193
|
|
The role of structural disorder in the rewiring of protein interactions through evolution,
Mol Cell Proteomics,
2012, Pubmed: 22389433,
DOI: 10.1074/mcp.M111.014969
|
|
A Novel Framework for the Comparative Analysis of Biological Networks,
PLoS ONE,
2012, 7 (2), Pubmed: 22363585,
DOI: 10.1371/journal.pone.0031220
|
|
Recycling side-effects into clinical markers for drug repositioning,
Genome Medicine,
2012, 4:3, Pubmed: 22283977,
DOI: 10.1186/gm302
|
2011
|
PSICQUIC and PSISCORE: accessing and scoring molecular interactions,
Nature Methods,
2011, 8:528–529, Pubmed: 21716279,
DOI: 10.1038/nmeth.1637
|
|
A Systematic Study of the Energetics Involved in Structural Changes upon Association and Connectivity in Protein Interaction Networks,
Structure,
2011, 19(6):881-9, Pubmed: 21645858,
DOI: 10.1016/j.str.2011.03.009,
Supplementary material
|
|
On the analysis of protein-protein interactions via knowledge-based potentials for the prediction of protein-protein docking,
Protein Sci,
2011, 20(3):529–541, Pubmed: 21432933,
DOI: 10.1002/pro.585
|
|
Three-dimensional modeling of protein interactions and complexes is going 'omics,
Curr Opin Struct Biol,
2011, 21(2):200-8, Pubmed: 21320770,
DOI: 10.1016/j.sbi.2011.01.005
|
|
Interactome mapping suggests new mechanistic details underlying Alzheimer's disease,
Genome Res,
2011, 21(3):364-76, Pubmed: 21163940,
DOI: 10.1101/gr.114280.110,
Supplementary material
|
|
Phospho3D 2.0: an enhanced database of three-dimensional structures of phosphorylation sites,
Nucleic Acids Res,
2011, 39(Database issue):D268-71, Pubmed: 20965970,
DOI: 10.1093/nar/gkq936
|
|
3did: identification and classification of domain-based interactions of known three-dimensional structure,
Nucleic Acids Res,
2011, 39(Database issue):D718–D723, Pubmed: 20965963,
DOI: 10.1093/nar/gkq962
|
2010
|
Revealing the molecular relationship between type 2 diabetes and the metabolic changes induced by a very-low-carbohydrate low-fat ketogenic diet,
Nutr Metab (Lond),
2010, 9(7):88, Pubmed: 21143928,
DOI: 10.1186/1743-7075-7-88
|
|
Uncovering new substrates for Aurora A kinase,
|
|
Systematic bioinformatics and experimental validation of yeast complexes reduces the rate of attrition during structural investigations,
Structure,
2010, 9(8):1075-1082, Pubmed: 20826334,
DOI: 10.1016/j.str.2010.08.001,
Supplementary material
|
|
Novel peptide-mediated interactions derived from high-resolution 3-dimensional structures,
PLoS Comput Biol,
2010, 6(5):e1000789, Pubmed: 20502673,
DOI: 10.1371/journal.pcbi.1000789
|
|
Structure of the FoxM1 DNA-recognition domain bound to a promoter sequence,
Nucleic Acids Res,
2010, 38(13):4527-38, Pubmed: 20360045,
DOI: 10.1093/nar/gkq194
|
|
Predicting protein-protein interaction specificity through the integration of three-dimensional structural information and the evolutionary record of protein domains,
Mol Biosyst,
2010, 6(4):741-9, Pubmed: 20237652,
DOI: 10.1039/B918395G
|
|
Unveiling the role of network and systems biology in drug discovery,
Trends in Pharmacological Sciences,
2010, 31(3):115-123, Pubmed: 20117850,
DOI: 10.1016/j.tips.2009.11.006
|
2009
|
Splitting statistical potentials into meaningful scoring functions: testing the prediction of near-native structures from decoy conformations,
BMC Struct Biol,
2009, 16(9):71, Pubmed: 19917096,
DOI: 10.1186/1472-6807-9-71
|
|
Pushing structural information into the yeast interactome by high-throughput protein docking experiments,
PLoS computational biology,
2009, 5(8):e1000490, Pubmed: 19714207,
DOI: 10.1371/journal.pcbi.1000490,
Supplementary material
|
|
Dynamic interactions of proteins in complex networks: a more structured view,
FEBS J,
2009, 276(19):5390-405, Pubmed: 19712106,
DOI: 10.1111/j.1742-4658.2009.07251.x
|
|
Exploiting gene deletion fitness effects in yeast to understand the modular architecture of protein complexes under different growth conditions,
BMC Systems Biology,
2009, 3:74, Pubmed: 19615085,
DOI: 10.1186/1752-0509-3-74
|
|
A network medicine approach to human disease,
FEBS Letters,
2009, 583(11):1759-65, Pubmed: 19269289,
DOI: doi:10.1016/j.febslet.2009.03.001
|
|
3did Update: domain-domain and peptide-mediated interactions of known 3D structure,
Nucleic Acids Res,
2009, 37(Database issue):D300-4, Pubmed: 18953040,
DOI: 10.1093/nar/gkn690
|
2008
|
Targeting and tinkering with interaction networks,
Nat Chem Biol,
2008, 4(11):666-73, Pubmed: 18936751,
DOI: 10.1038/nchembio.119
|
|
Approved drug mimics of short peptide ligands from protein interaction motifs,
J Chem Inf Model,
2008, 48(10):1943-8, Pubmed: 18826301,
DOI: 10.1021/ci800174c
|
|
Contextual specificity in peptide-mediated protein interactions,
PLoS One,
2008, 3(7):e2524, Pubmed: 18596940,
DOI: 10.1371/journal.pone.0002524
|
|
Incorporating high-throughput proteomics experiments into structural biology pipelines: Identification of the low-hanging fruits,
Proteomics,
2008, 8(10):1959–1964, Pubmed: 18491310,
DOI: 10.1002/pmic.200700966
|
|
Towards a molecular characterisation of pathological pathways,
FEBS Lett,
2008, 582(8):1259-65, Pubmed: 18282477,
DOI: 10.1016/j.febslet.2008.02.014
|
|
A molecular interpretation of genetic interactions in yeast,
FEBS Lett,
2008, 582(8):1245-50, Pubmed: 18282475,
DOI: 10.1016/j.febslet.2008.02.020
|
2007
|
Shaping the future of interactome networks,
Genome Biol,
2007, 8(10):316, Pubmed: 17980052,
DOI: 10.1186/gb-2007-8-10-316
|