Our webservers and other resources
|
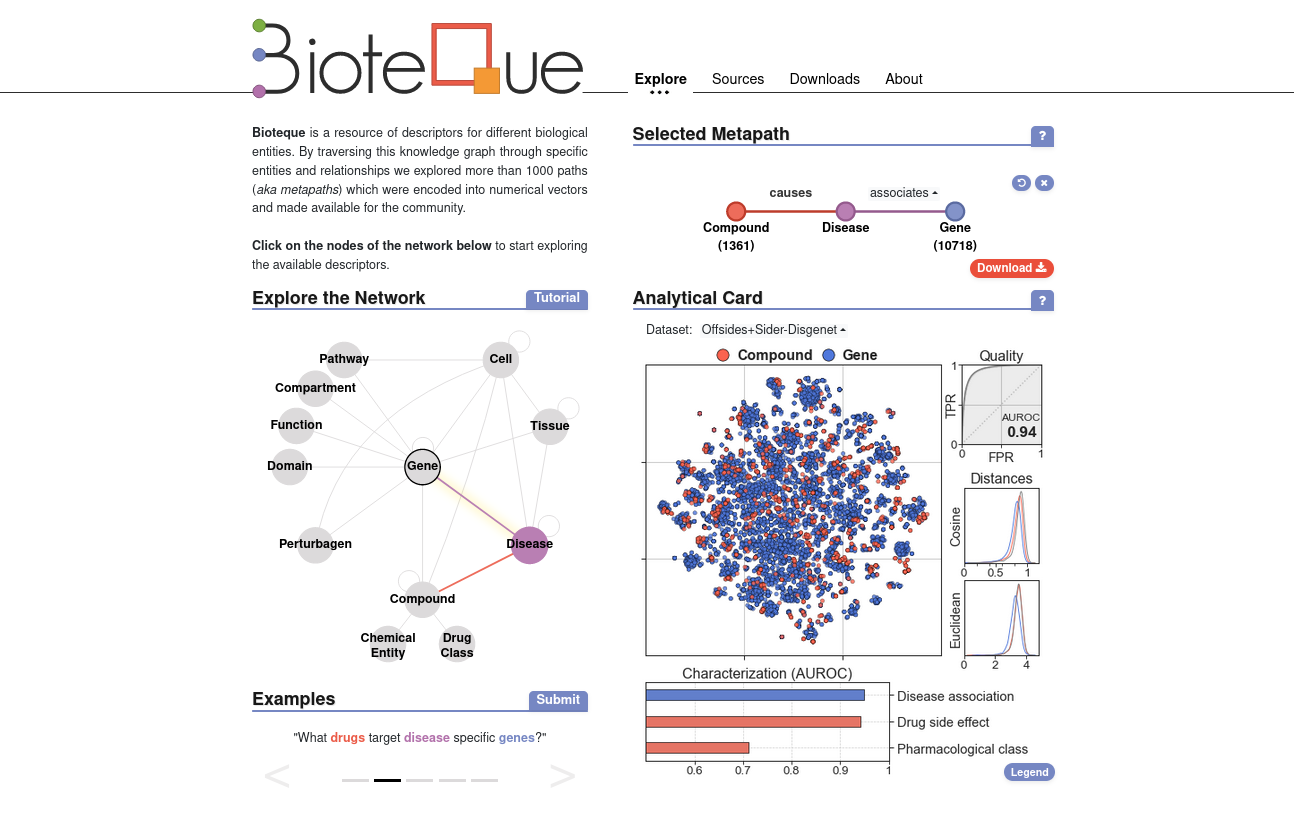
BiotequeThe Bioteque is a resource of descriptors for different biological entities. By traversing this knowledge graph through specific entities and relationships we explored more than 1000 paths (aka metapaths) which were encoded into numerical vectors and made available for the community. You can access the server at https://bioteque.irbbarcelona.org/. |
|
|
|
|
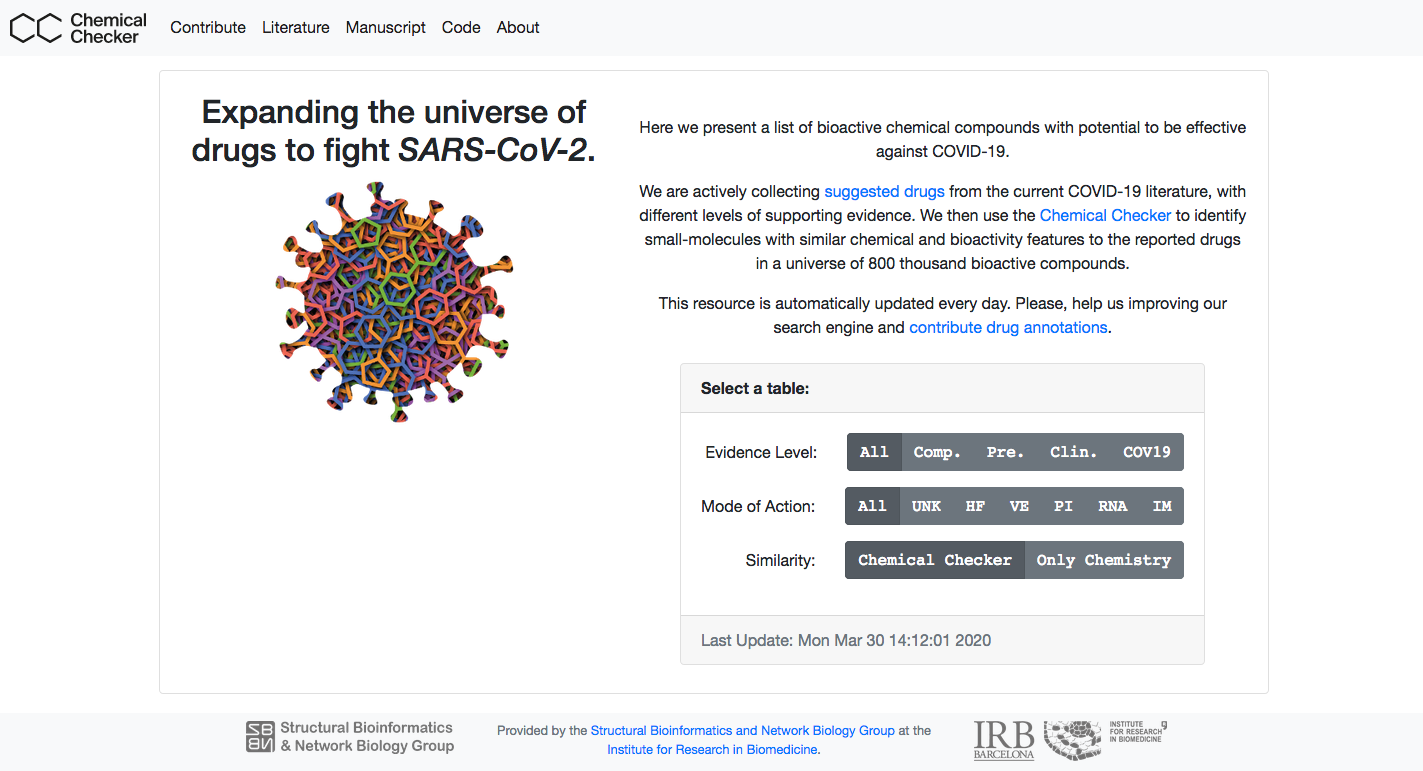
Expanding the universe of drugs to fight SARS-CoV-2Here we present a list of bioactive chemical compounds with potential to be effective against COVID-19. We are actively collecting suggested drugs from the current COVID-19 literature, with different levels of supporting evidence. We then use the Chemical Checker to identify small-molecules with similar chemical and bioactivity features to the reported drugs in a universe of 800 thousand bioactive compounds. This resource is automatically updated every day. Please, help us improving our search engine and contribute drug annotations. You can access the server at https://sbnb.irbbarcelona.org/covid19/. |
|
|
|
|
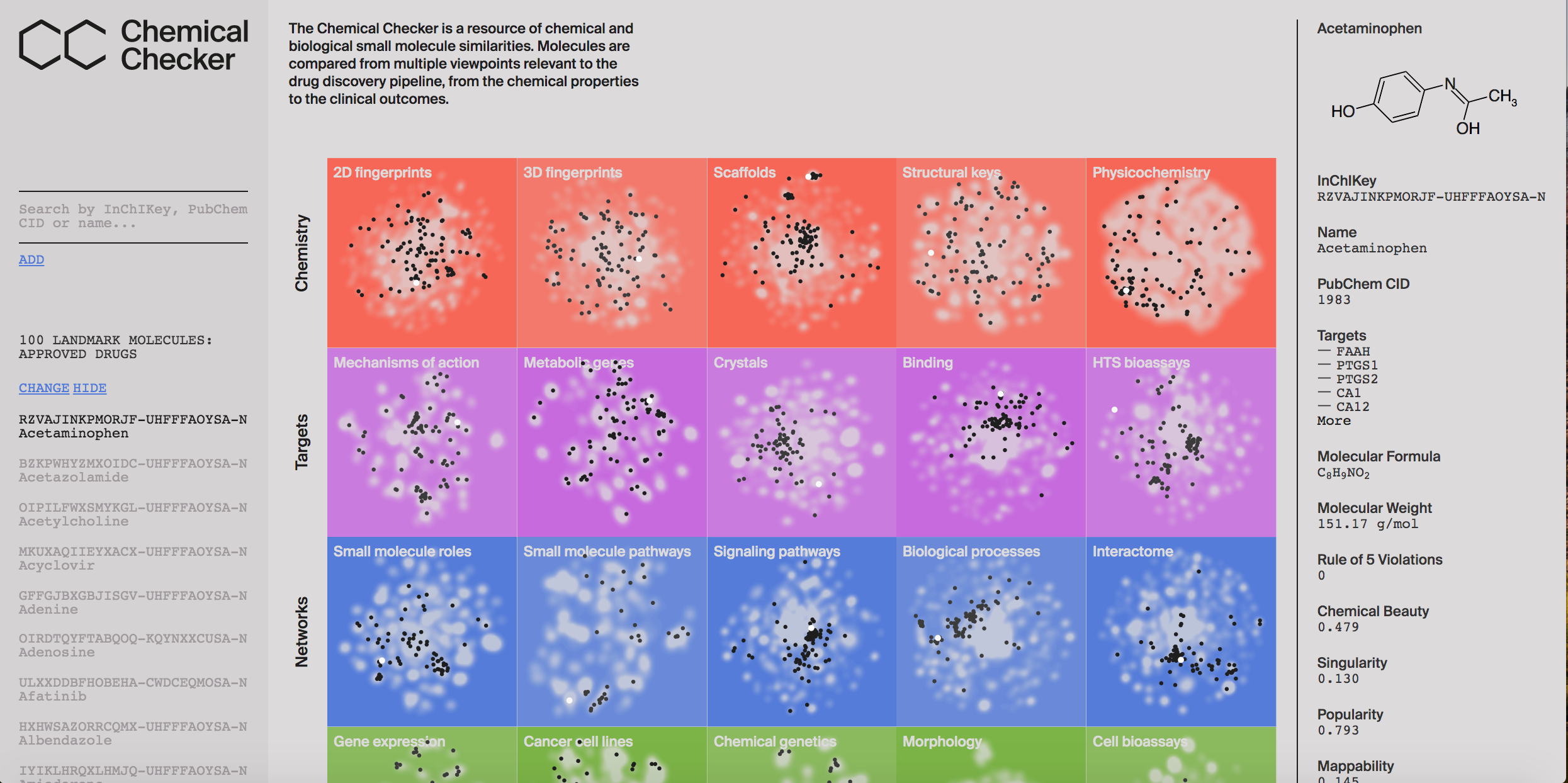
Chemical CheckerThe Chemical Checker (CC) is a resource that provides processed, harmonized and integrated bioactivity data on 800,000 small molecules. The CC divides data into five levels of increasing complexity, ranging from the chemical properties of compounds to their clinical outcomes. In between, it considers targets, off-targets, perturbed biological networks and several cell-based assays such as gene expression, growth inhibition and morphological profilings. In the CC, bioactivity data are expressed in a vector format, which naturally extends the notion of chemical similarity between compounds to similarities between bioactivity signatures of different kinds. You can access the server at https://chemicalchecker.com/. |
|
|
|
|
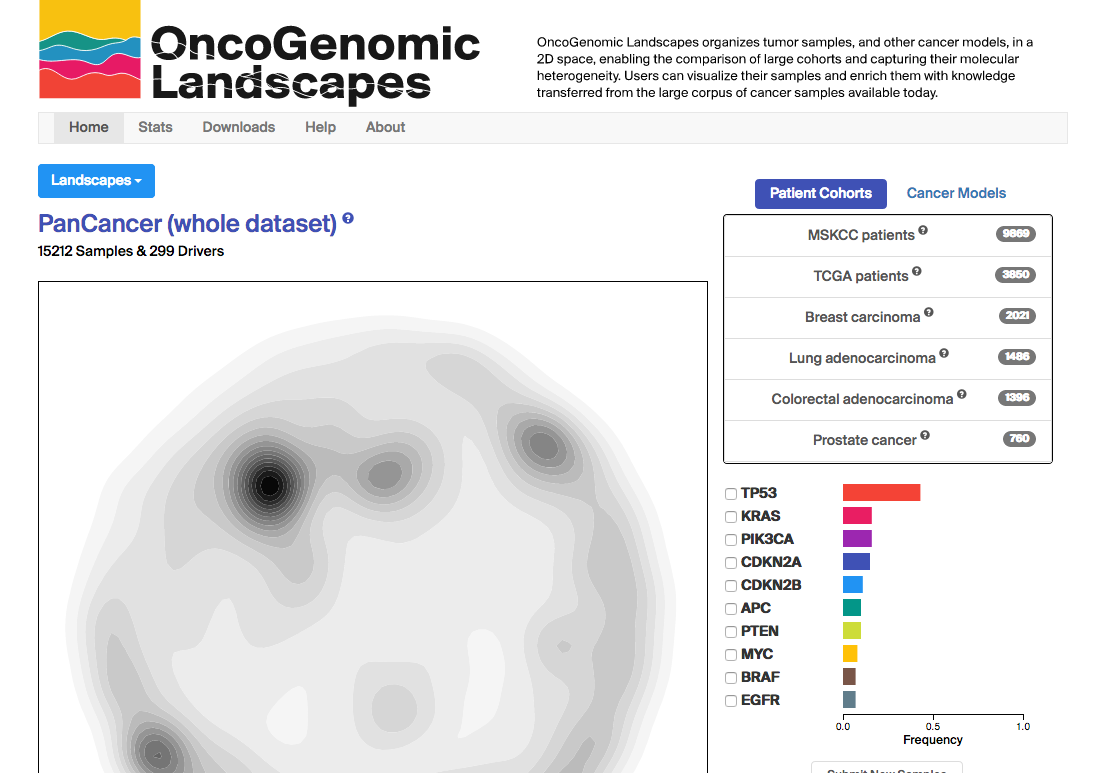
OncoGenomic LandscapesOncoGenomic Landscapes is a visualization tool that organizes tumor samples, and other cancer models, in a 2D space, enabling the comparison of large cohorts and capturing their molecular heterogeneity. Our resource includes information on the main cohorts published to date and, in addition, offers the possibility of mapping new samples and cohorts, providing an intuitive means to visualize user data and enrich it with knowledge transferred from the large corpus of cancer samples available. You can access the server at https://oglandscapes.irbbarcelona.org/. |
|
|
|

|
Cancer PanorOmicsCancer PanorOmics is a web-based resource to contextualize genomic variations detected in a personal cancer genome within the body of clinical and scientific evidence available for 26 tumor types, offering complementary cohort- and patient-centric views. Additionally, it explores the cellular environment of mutations by mapping them on the human interactome and providing quasi-atomic structural details, whenever available. You can access the server at http://panoromics.irbbarcelona.org. |
|
|
|
|
DynBench3DDynBench3D is a web-based resource that allows the automated generation of benchmark sets, composed of target and template protein complexes of known 3D structure, through a user friendly interface. Users can tune redundancy reduction parameters, interactively select and inspect targets based on structural determination method, complex architecture, or depending on templates availability and download the benchmark. https://dynbench3d.irbbarcelona.org/. |
|
|
|

|
dSysMapdSysMap is a resource for the systematic mapping of disease-related missense mutations on the human structural interactome. It summarizes mutational data in a systemic context, and provides a unique tool to the biologist to easily grab the molecular relationships between diseases and formulate hypotheses about their mechanism of action. In addition to the precompiled data for thousands of diseases in OMIM, dSysMap allows to upload newly discovered mutations so that disease context can be readily incorporated into primary sequencing studies. You can access the server at http://dsysmap.irbbarcelona.org. |
|
|
|

|
IntSideIntSide is a web server to elucidate the molecular processes involved in drug side effects through the integration of chemistry and biology. IntSide currently catalogues 1,175 side effects caused by 996 drugs, associated with drug features divided into eight categories, belonging to either biology or chemistry. On the biological side, IntSide reports drug targets and off-targets, pathways, molecular functions and biological processes. From a chemical viewpoint, it includes molecular fingerprints, scaffolds and chemical entities. Finally, we also integrate additional biological data, such as protein interactions and disease-related genes, to facilitate mechanistic interpretations. IntSide is available at http://intside.irbbarcelona.org. |
|
|
|

|
Interactome3DInteractome3D is a web service for the structural annotation of protein-protein interaction networks. You can submit your interactions and the server will find all the available structural data for both the single interactors and the interactions themselves. Additionally you can also visualize and download structural information for interactions involving a set of proteins or interactomes for several model organisms. You can access the server at http://interactome3d.irbbarcelona.org. |
|
|
|

|
3didThe database of three-dimensional interacting domains (3did) is a collection of protein interactions for which high-resolution three-dimensional structures are known. 3did exploits the a availability of structural data to provide molecular details on interactions between two globular domains as well as novel domain–peptide interactions, derived using a recently published method from our lab. The interface residues are presented for each interaction type individually, plus global domain interfaces at which one or more partners (domains or peptides) bind. The 3did web server at http://3did.irbbarcelona.org visualizes these interfaces along with atomic details of individual interactions using Jmol. The complete contents are also available for download. |
 |
NetAlignerThe complex nature of biological systems requires intricate networks of protein-protein interactions that coordinate macromolecular assemblies and pathways to fulfil particular tasks in a cell. We developed NetAligner, a novel network alignment tool that allows the identification of conserved protein complexes and pathways across organisms and thus the study of how those interaction networks evolved. NetAligner is able to detect potential false positives by considering protein sequence similarities and interaction reliabilities, and addresses the issue of missing relationships through the prediction of likely conserved interactions. Gaps and mismatches in the alignment account for small amounts of network rewiring during evolution, and alignment solutions are tested for statistical significance. The web server implementation of the NetAligner algorithm features complex, pathway and interactome to interactome alignment for seven model organisms, H.sapiens, M. musculus, D.melanogaster, C. elegans, A.thaliana, S.cerevisiae and E. coli. The user can specify query complexes and pathways of arbitrary topology for alignment against a target interactome selected from our network database or choose two species interactomes to search for conserved complexes and subnetworks. Alignment solutions can be downloaded or directly visualized in the browser. The software package can be downloaded for Unix/Linux/Mac and Windows. |
