Encircling the regions of the pharmacogenomic landscape that determine drug response
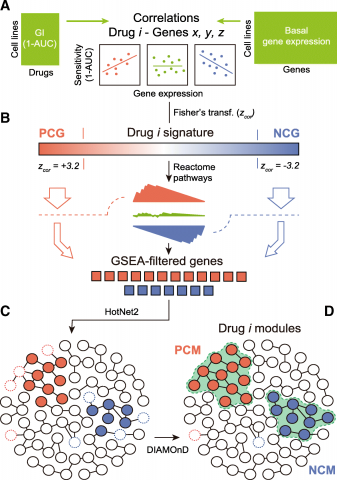
Fernández-Torras A, Duran-Frigola M, Aloy P, Background:The integration of large-scale drug sensitivity screens and genome-wide experiments is changing thefield of pharmacogenomics, revealing molecular determinants of drug response without the need for previousknowledge about drug action. In particular, transcriptional signatures of drug sensitivity may guide drugrepositioning, prioritize drug combinations, and point to new therapeutic biomarkers. However, the inherentcomplexity of transcriptional signatures, with thousands of differentially expressed genes, makes them hard tointerpret, thus giving poor mechanistic insights and hampering translation to clinics. Methods:To simplify drug signatures, we have developed a network-based methodology to identify functionallycoherent gene modules. Our strategy starts with the calculation of drug-gene correlations and is followed by apathway-oriented filtering and a network-diffusion analysis across the interactome. Results:We apply our approach to 189 drugs tested in 671 cancer cell lines and observe a connection betweengene expression levels of the modules and mechanisms of action of the drugs. Further, we characterize multipleaspects of the modules, including their functional categories, tissue-specificity, and prevalence in clinics. Finally, weprove the predictive capability of the modules and demonstrate how they can be used as gene sets inconventional enrichment analyses. Conclusions:Network biology strategies like module detection are able to digest the outcome of large-scalepharmacogenomic initiatives, thereby contributing to their interpretability and improving the characterization of thedrugs screened.
Genome Medicine,
2019, 11, 17
Pubmed: 30914058
Direct link: https://doi.org/10.1186/s13073-019-0626-x
