dSysMap: Exploring the edgetic role of disease mutations
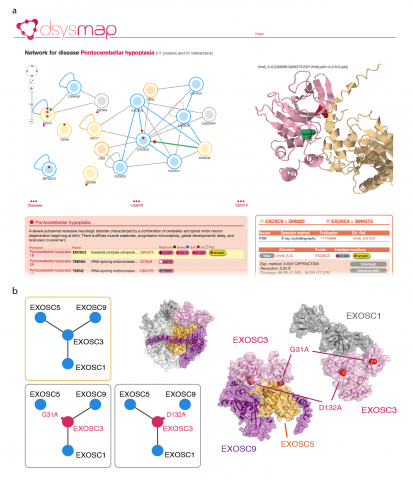
Mosca R, Tenorio-Laranga J, Olivella R, Alcalde V, Ceol A, Soler-López M, Aloy P, Considering pathological genetic variants within the context of the human interactome network can help understanding the intricate genotype-to-phenotype relationships behind human diseases. It allows, for instance, to distinguish between changes that totally suppress a gene product (i.e. node removal) from the ones that might affect only one of its functions, modulating the way in which the protein interacts with its partners (i.e. edge-specific or edgetic1). Here, we present dSysMap, a resource for the systematic mapping of disease-related missense mutations on the structurally annotated binary human interactome from Interactome3D.
Nature Methods,
2015, 12, 167–168
Pubmed: 25719824
Direct link: 10.1038/nmeth.3289
