Conditional Epistatic Interaction Maps Reveal Global Functional Rewiring of Genome Integrity Pathways in Escherichia coli
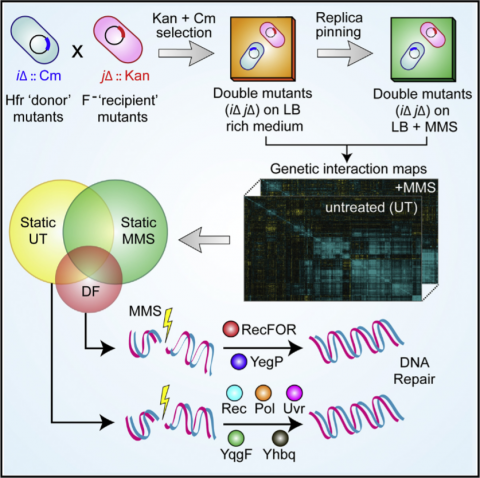
Kumar A, Beloglazova N, Bundalovic-Torma C, Phanse S, Deineko V, Gagarinova A, Musso G, Vlasblom J, Lemak S, Hooshyar M, Minic Z, Wagih O, Mosca R, Aloy P, Golshani A, Parkinson J, Emili A, F. Yakunin A, Babu M, As antibiotic resistance is increasingly becoming a public health concern, an improved understanding of the bacterial DNA damage response (DDR), which is commonly targeted by antibiotics, could be of tremendous therapeutic value. Although the genetic components of the bacterial DDR have been studied extensively in isolation, how the underlying biological pathways interact functionally remains unclear. Here, we address this by performing systematic, unbiased, quantitative synthetic genetic interaction (GI) screens and uncover widespread changes in the GI network of the entire genomic integrity apparatus of Escherichia coli under standard and DNA-damaging growth conditions. The GI patterns of untreated cultures implicated two previously uncharacterized proteins (YhbQ and YqgF) as nucleases, whereas reorganization of the GI network after DNA damage revealed DDR roles for both annotated and uncharacterized genes. Analyses of pan-bacterial conservation patterns suggest that DDR mechanisms and functional relationships are near universal, highlighting a modular and highly adaptive genomic stress response
Cell Reports,
2016, 14, 648-661
Pubmed: 26774489
Direct link: 10.1016/j.celrep.2015.12.060
